Abstract
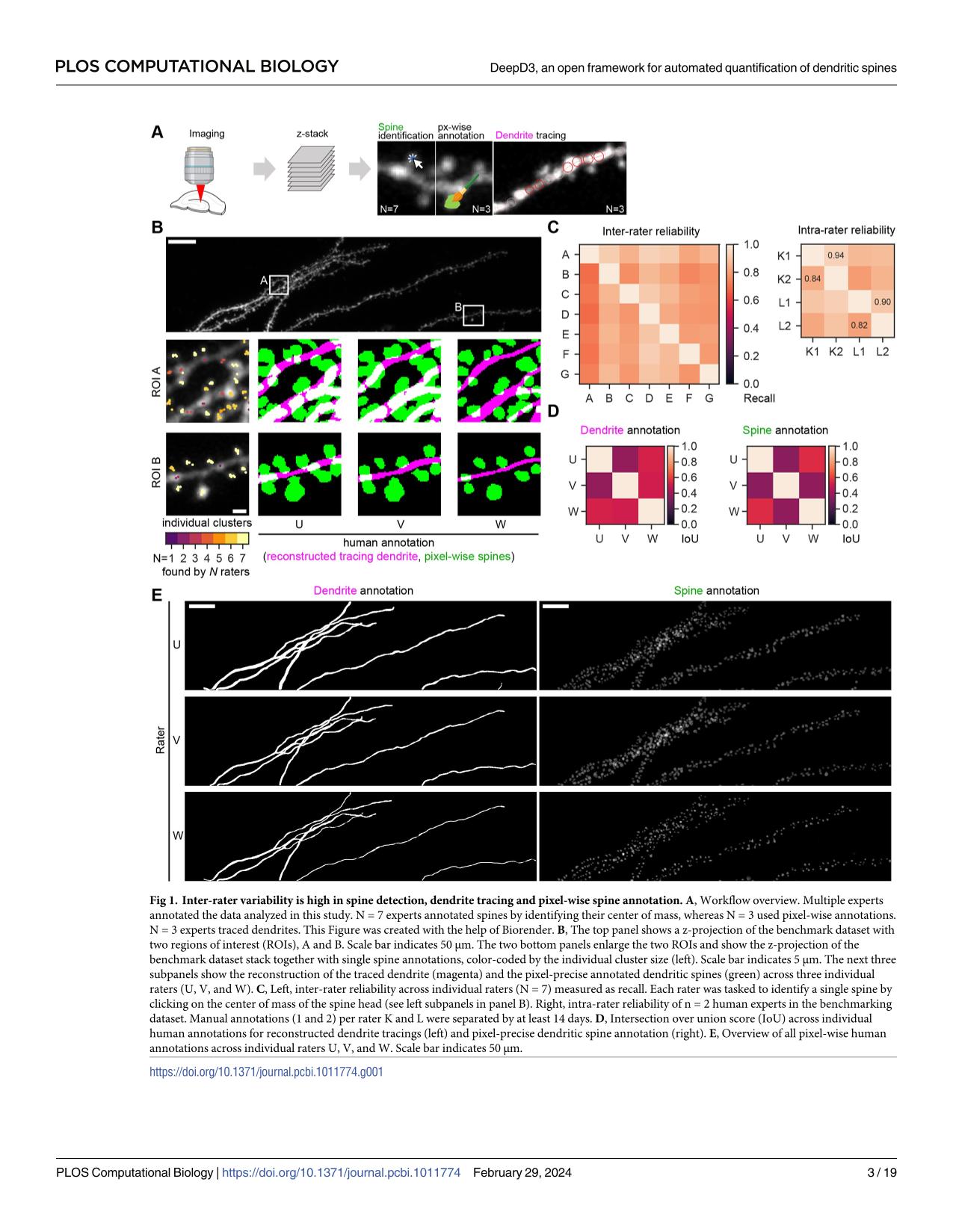
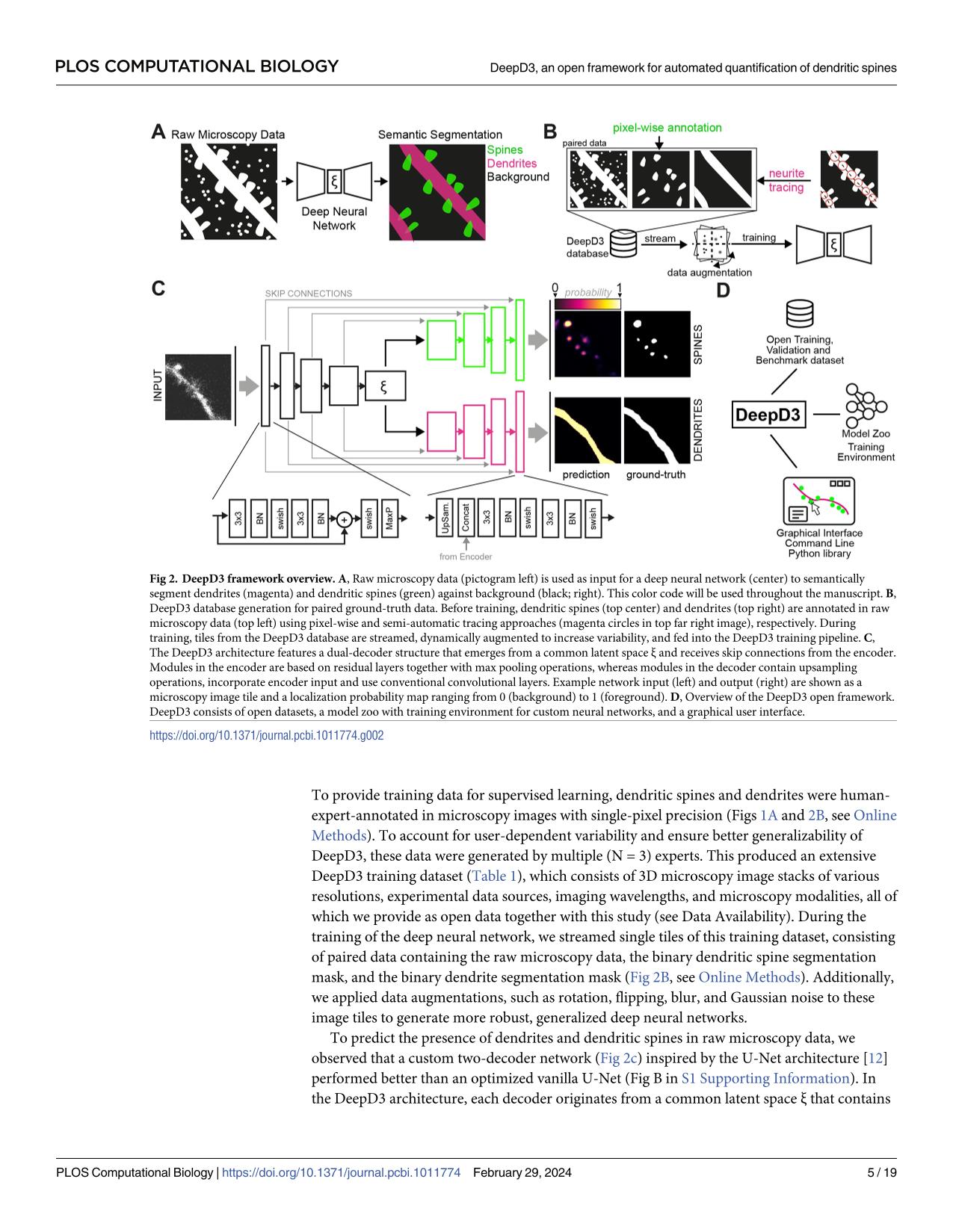
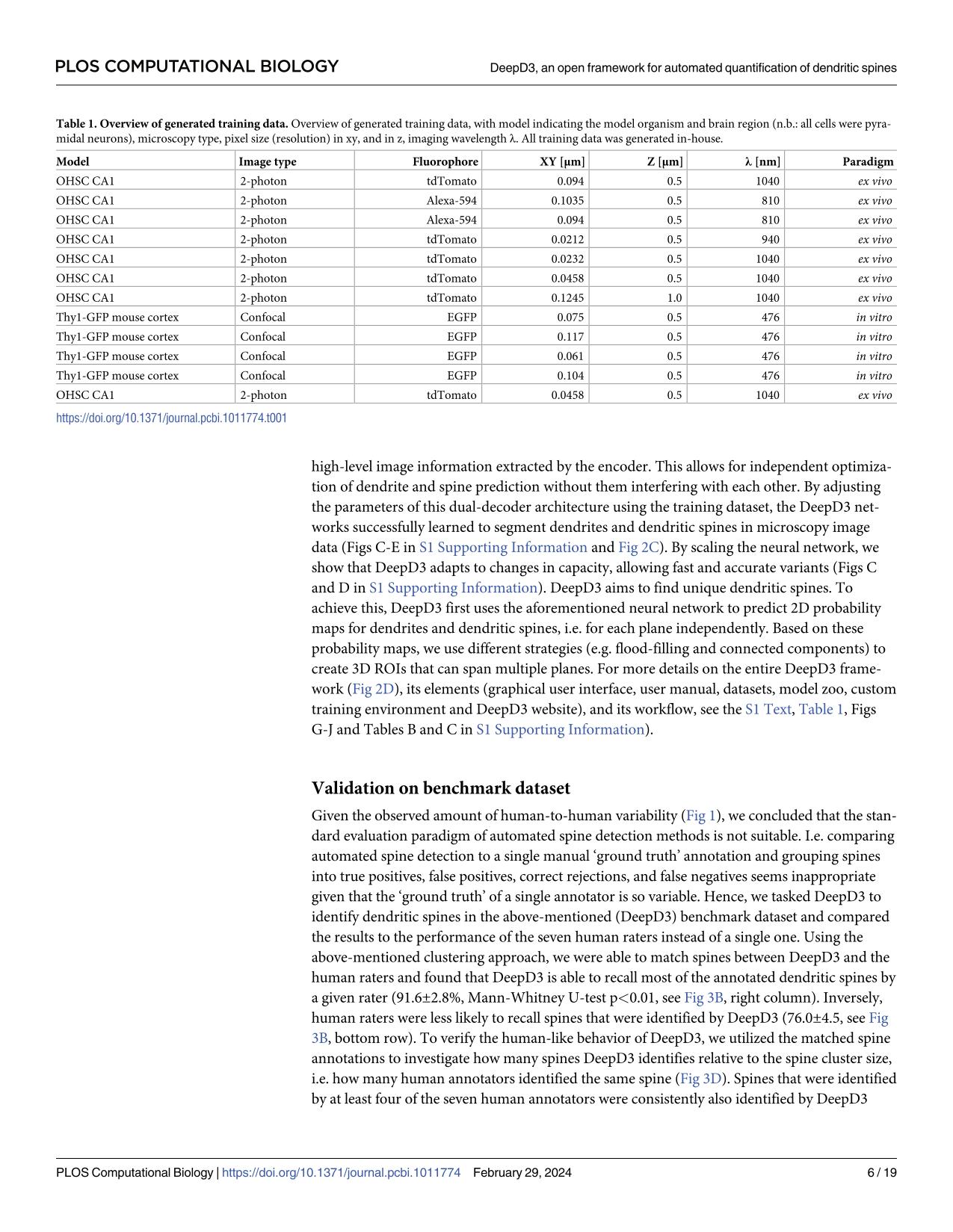
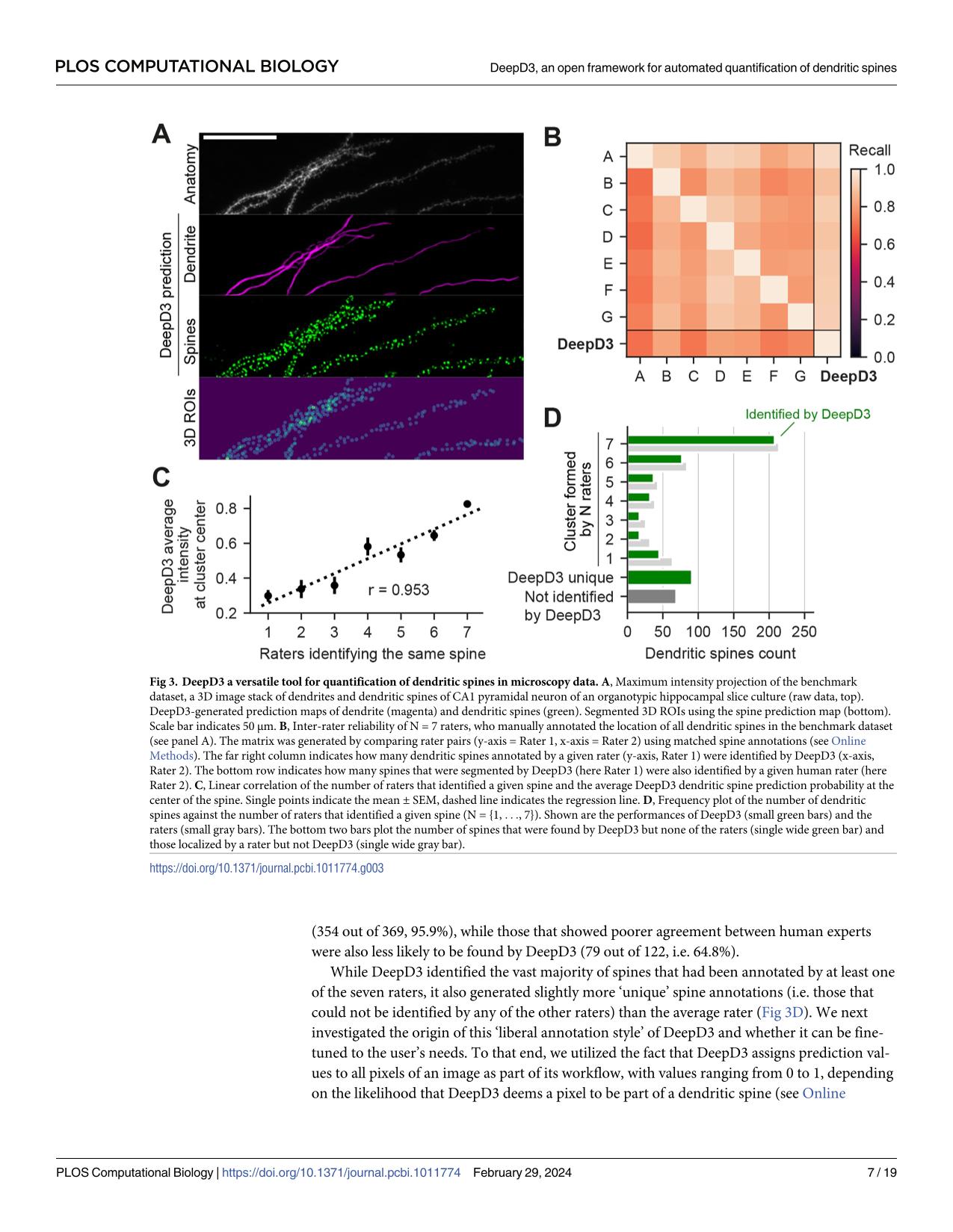
Dendritic spines are the seat of most excitatory synapses in the brain, and a cellular structure considered central to learning, memory, and activity-dependent plasticity. The quantification of dendritic spines from light microscopy data is usually performed by humans in a painstaking and error-prone process. We found that human-to-human variability is substantial (inter-rater reliability 82.2±6.4 %), raising concerns about the reproducibility of experiments and the validity of using human-annotated 'ground truth' as an evaluation method for computational approaches of spine identification. To address this, we present DeepD3, an open deep learning-based framework to robustly quantify dendritic spines in microscopy data in a fully automated fashion. DeepD3's neural networks have been trained on data from different sources and experimental conditions, annotated and segmented by multiple experts and they offer precise quantification of dendrites and dendritic spines. Importantly, these networks were validated in a number of datasets on varying acquisition modalities, species, anatomical locations and fluorescent indicators. The entire DeepD3 open framework, including the fully segmented training data, a benchmark that multiple experts have annotated, and the DeepD3 model zoo is fully available, addressing the lack of openly available datasets of dendritic spines while offering a ready-to-use, flexible, transparent, and reproducible spine quantification method.
How DeepD3 works
DeepD3 is installable using pip install deepd3,
see the Code section for further instructions.
The most convenient way is to use our graphical user interface (GUI)
to open your data and process it using our pipeline. Pre-trained
deep neural networks for dendritic spines and dendrite segmentation
can be found in our Model Zoo.
Tutorial: DeepD3 inference
Tutorial: DeepD3 train your own neural network
If you trained your network with a fixed shape (e.g. 128x128x1), you can use this script/Jupyter notebook to convert your network to a generic one.
Frequently Asked Questions NEW!
- I get the following error in Colab:
AttributeError: module 'keras.utils.generic_utils' has no attribute 'get_custom_objects'
Link to solution
The DeepD3 benchmark dataset
You are able to download all DeepD3 benchmark dataset related files on Zenodo. For your convenience, we provide also the two-photon microscopy stack here.
3D preview
Model Zoo
We provide openly our pre-trained model zoo on Zenodo and on our respective Model Zoo section here. If you would like to contribute your custom model, please use the following form to submit your model.
Models trained on DeepD3 Training Dataset with DeepD3 Architecture
- DeepD3_8F.h5 - 8 base filters, original resolution
- DeepD3_16F.h5 - 16 base filters, original resolution
- DeepD3_32F.h5 - 32 base filters, original resolution
- DeepD3_8F_94nm.h5 - 8 base filters, resized to 94 nm xy resolution
- DeepD3_16F_94nm.h5 - 16 base filters, resized to 94 nm xy resolution
- DeepD3_32F_94nm.h5 - 32 base filters, resized to 94 nm xy resolution
Paper
published in PLOS computational biology
Cite DeepD3
Article Source:
DeepD3, an open framework for automated quantification of dendritic spines
Fernholz MHP,
Guggiana Nilo DA,
Bonhoeffer T,
Kist AM
(2024)
DeepD3, an open framework for automated quantification of dendritic spines.
PLOS Computational Biology 20(2): e1011774.
https://doi.org/10.1371/journal.pcbi.1011774
@article{10.1371/journal.pcbi.1011774, doi = {10.1371/journal.pcbi.1011774}, author = {Fernholz, Martin H. P. AND Guggiana Nilo, Drago A. AND Bonhoeffer, Tobias AND Kist, Andreas M.}, journal = {PLOS Computational Biology}, publisher = {Public Library of Science}, title = {DeepD3, an open framework for automated quantification of dendritic spines}, year = {2024}, month = {02}, volume = {20}, url = {https://doi.org/10.1371/journal.pcbi.1011774}, pages = {1-19}, number = {2}, }